Paneles TruSight. Genética y Oncología

The NGS Clinical Exome Solution (CES v3) of SOPHiA GENETICS covers the coding regions (± 5 bp of intronic regions) of 4,728 genes, the entire mitochondrial genome and ~200 non-coding variants with known pathogenicity in deep introns/enhancer/promoter genes. This new and improved probe design of the v3 provides increased capacity for detection of rare and inherited disorders.
With the clinical exome study for clinical diagnosis, mutations in genes not related to the clinical phenotype of the patient but with clinical relevance can be detected. Exome sequencing thus helps early detection and early treatment of this genetic diseases.
CES v3 offers a streamlined end-to-end workflow (from sample to variant report), making it faster and easier to assessment of challenging Mendelian disorders, freeing up time and resources.
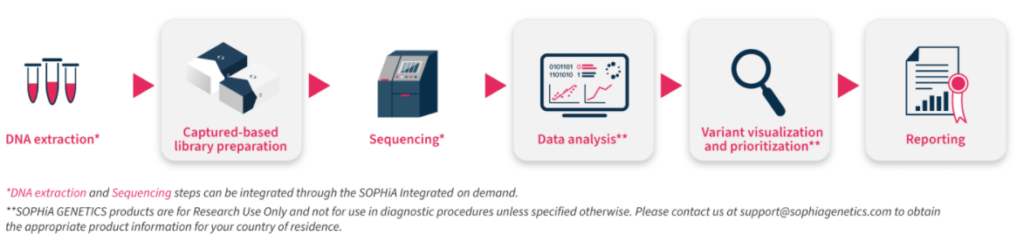
This NGS solution includes a capture-based target enrichment kit, as well as the functionality and analytical capabilities of the SOPHiA DDM™ platform for data interpretation.
The SOPHiA DDM™ platform features intuitive variant filters and prioritization options to streamline the interpretation process and help you greatly reduce turnaround time.
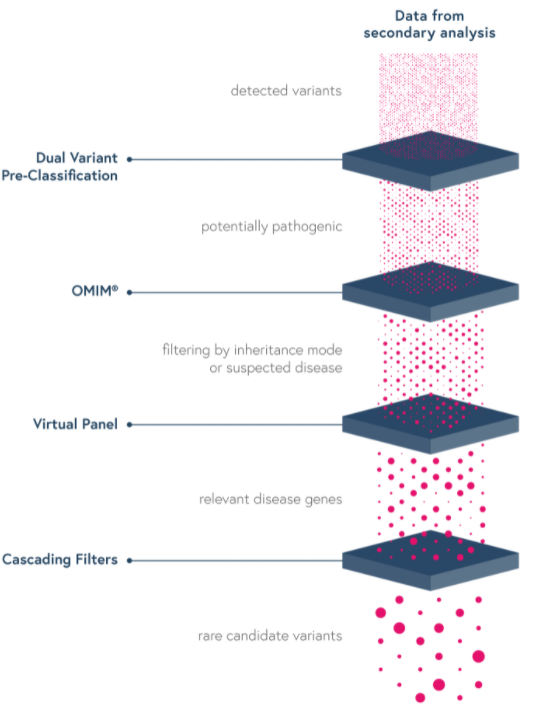
Through SOPHiA DDM™, you can also have access to Alamut Visual Plus, a full-genome browser that integrates numerous curated genomic and literature databases, guidelines, missense and slicing predictors, thus enabling a deeper variant exploration.
In addition to the Clinical Exome kit, we have other more specific panels for clinical diagnosis in areas such as metabolism, oncology, genetics or cardiology.
| Presentación |
|---|
| 16 test |
| 32 test |
| 48 test |
Public Documentation
2021 © Diagnóstica Longwood SL
This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.
Strictly Necessary Cookie should be enabled at all times so that we can save your preferences for cookie settings.
If you disable this cookie, we will not be able to save your preferences. This means that every time you visit this website you will need to enable or disable cookies again.
This website uses Google Analytics to collect anonymous information such as the number of visitors to the site, and the most popular pages.
Keeping this cookie enabled helps us to improve our website.
Please enable Strictly Necessary Cookies first so that we can save your preferences!
Más información sobre nuestra política de cookies
