The Oncodeep kit from OncoDNA offers a comprehensive solution for identifying mutations, variants, and fusions in the somatic line of genes associated with solid tumors through Next-Generation Sequencing (NGS).
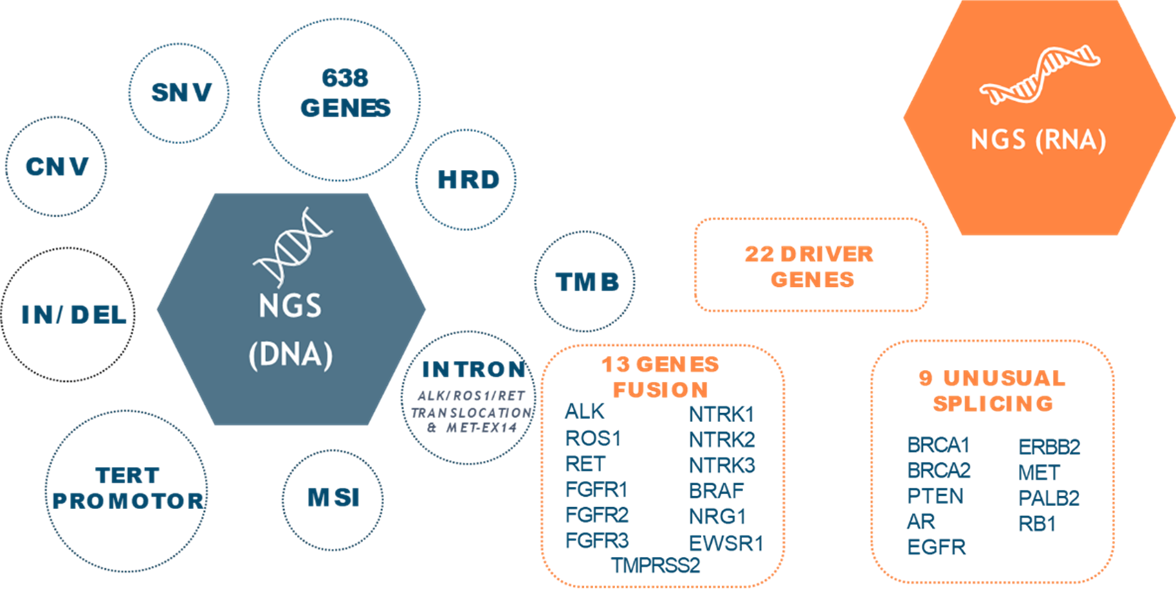
Designed by oncology experts, this kit includes the most relevant and complete gene panel in the field of cancer, consisting of 638 genes, which allows for coverage of all clinically relevant oncological targets and intronic translocations in genes such as ALK, ROS1, RET, and MET exon 14.
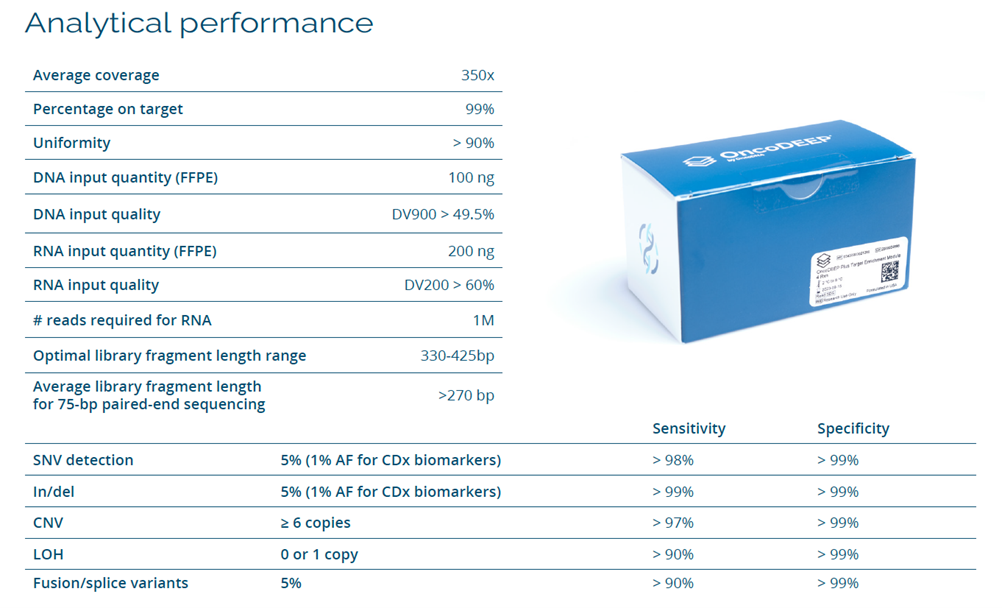
In addition to detecting SNVs, insertions, deletions, CNVs, and LOH, the kit also identifies complex genomic signatures such as HRD, MSI, and TMB, providing a valuable tool for cancer research and precise diagnosis.
It includes genes involved in Homologous Recombination Deficiency (HRD) for predicting response to PARP inhibitors. Additionally, it includes the detection of over 2200 SNPs for LOH analysis in key genes associated with immunotherapy response and in sub-telomeric regions to determine the HRD phenotype. It also detects fusions in 13 proven relevant genes and 9 unusual splicing variants from RNA.
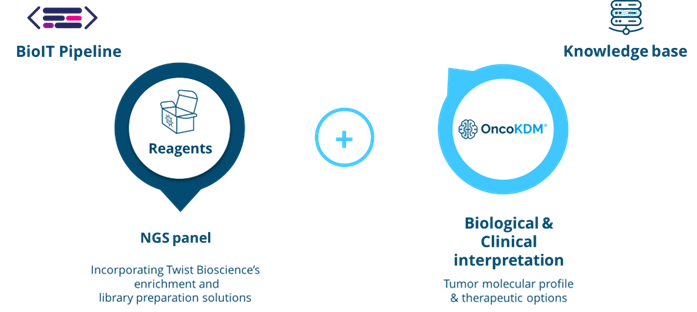
The offered kit provides a comprehensive solution by including library preparation reagents for sequencing over 600 cancer biomarkers. This offering is complemented by OncoDNA’s data analysis and clinical interpretation tools, called OncoKDM. This enables rapid analysis of a patient’s tumor molecular profile and the efficient generation of a detailed clinical report.
For which cases is it recommended?
The test is recommended for solid tumors in adults at stages 3-4, glioblastomas, pediatric tumors, and tumors of unknown primary cause. It can be performed at diagnosis, when first-line treatment is ineffective, in cases of recurrent tumors, very aggressive and/or rare tumors, and tumors of unknown primary cause.
From a tumor tissue sample, the OncoDEEP kit includes screening for genomic alterations (DNA and RNA) and genomic signatures that provide information on sensitivity and resistance to treatments such as targeted therapy, immunotherapy, hormone therapy, and chemotherapy, whether approved or in clinical trials.
Additionally, it is capable of detecting most actionable mutations.
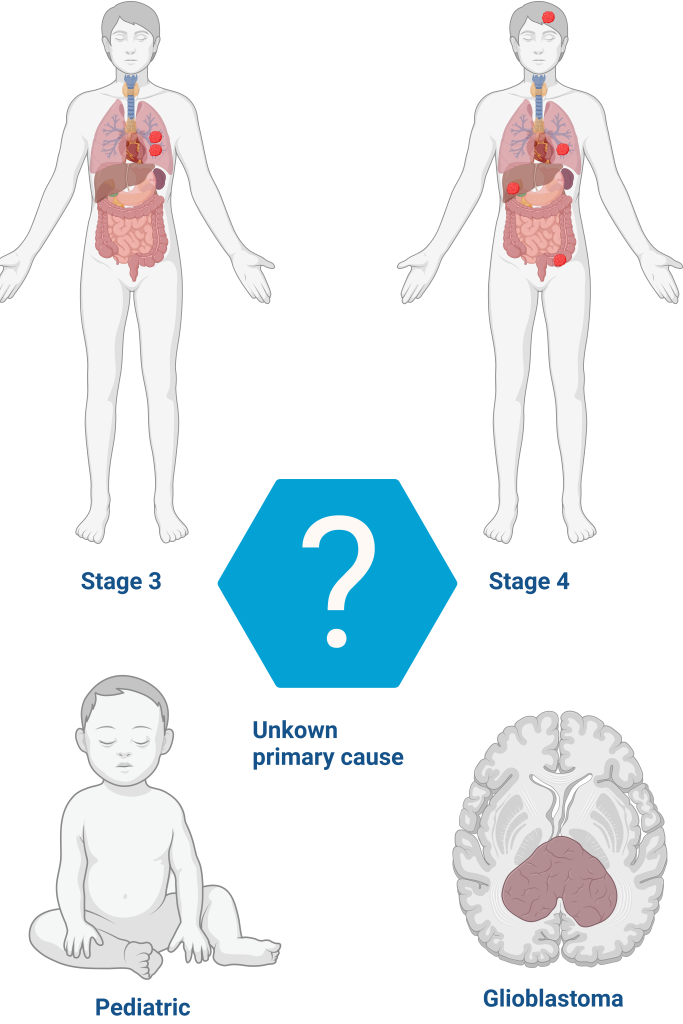
All¹ solid tumors
Tumor mutational burden (TMB): Pembrolizumab
Microsatellite instability (MSI): Pembrolizumab
NTRK: Entrectinib and Larotrectinib
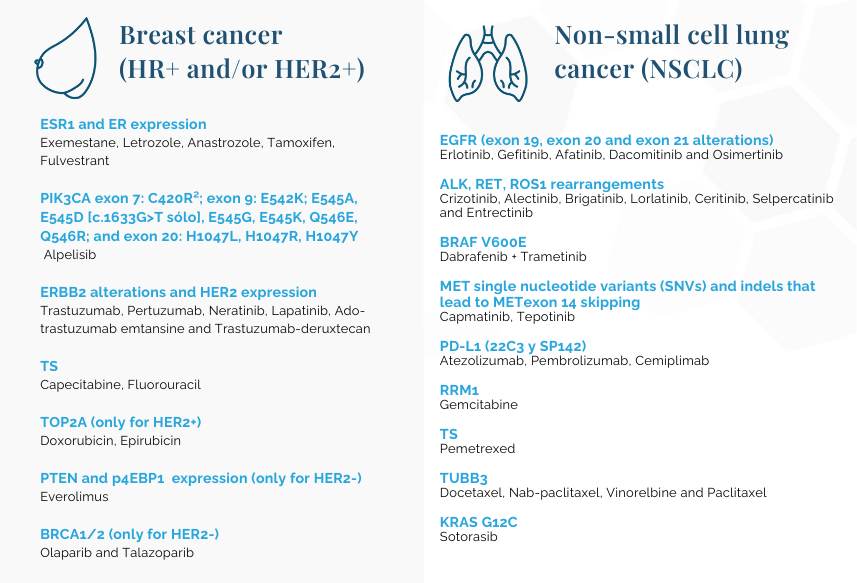

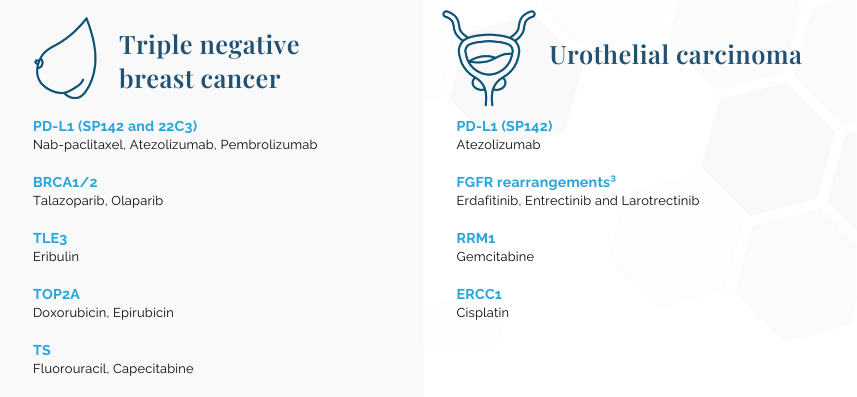
¹Tumor mutational burden (TMB) and microsatellite instability (MSI) scores will be calculated for any OncoDEEP order.
² DNA/RNA
³ Protein
PROTOCOL
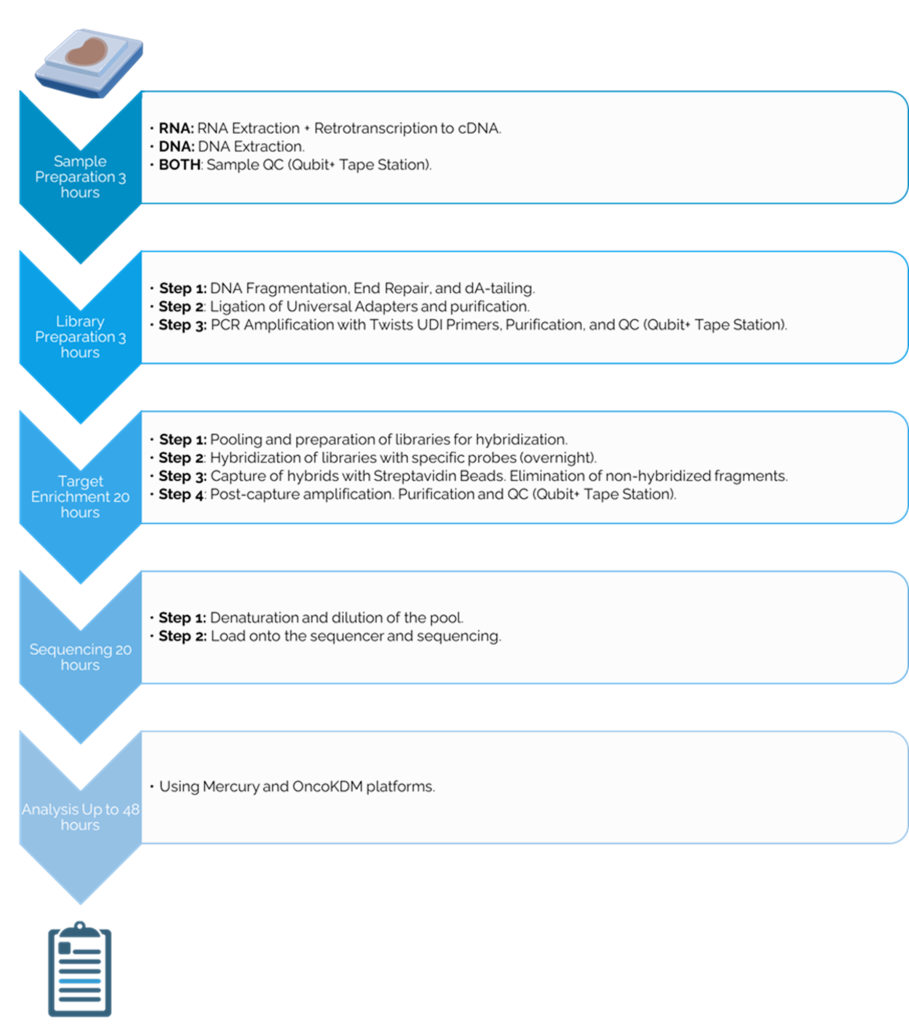
The offered solution is based on the use of a kit with reagents compatible with DNA and RNA extracted from paraffin-embedded tumor samples, with a recommended starting amount of 30-200 ng of DNA and 200-400 ng of RNA depending on the quality of the nucleic acids.
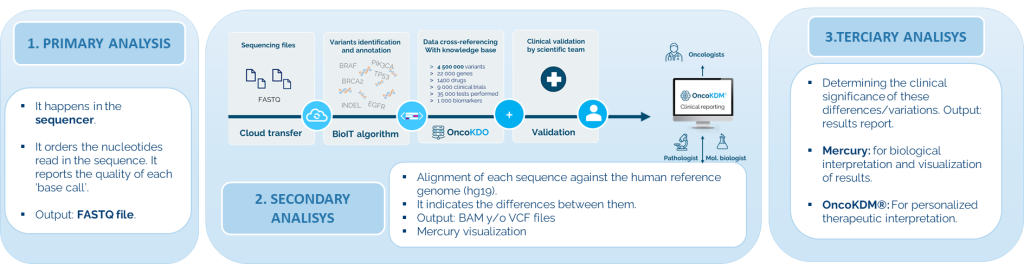
The DNA/RNA sequencing process of FFFPE tumor tissue begins with the extraction of samples, followed by library preparation and hybridization with capture probes using Twist technology. Subsequently, amplification and purification are carried out before sequencing on Illumina NextSeq or NovaSeq platforms. Finally, data analysis is performed using the OncoDEEP pipeline, followed by personalized therapeutic interpretation through OncoKDM.
SOFTWARE
The solution includes the OncoKDM® software for a detailed interpretation of results. This cloud-based platform converts NGS data into useful clinical information, allowing for easy visualization and review of reports. OncoKDM® can integrate NGS results with MSI, TMB, and HRD with IHC. The complete and interactive report includes updated information on approved treatments and research for each patient, along with NGS quality control data, clinical information of patients, and comprehensive annotations of NGS variants.
It is created by a company certified for ISO 13485 and ISO 27001, complying with Data Protection regulations. Data is stored on ISO 27001 certified servers, complying with GDPR (EU Regulation 2016/679).
The software can store FASTQ and BAM data for at least three years, and .vcf files indefinitely. Workflows and analysis are validated, generating “Quality Reports” for each sample. In addition to aligning sequences and identifying variants, it annotates and classifies variants based on their pathogenicity.
It integrates access to clinical and population databases, and updates somatic cancer data. It classifies variants according to their biological and therapeutic impact following ACMG/AMP guidelines. Besides predicting therapy sensitivity and offering information on clinical trials, it generates customizable reports manually reviewed and based on updated literature.
SPECIFICATIONS
The offered solution consists of a target region enrichment solution through double-stranded DNA probe capture technology by TWIST, ensuring sequencing with an average depth of >350X and coverage uniformity of >90%. It allows the detection of variants such as SNVs, indels, and CNVs with high sensitivity and specificity. Additionally, it identifies markers such as LOH and complex genomic signatures like HRD, MSI, and TMB. It also detects intronic translocations and exon skipping.
The included software complies with quality and data protection regulations, enables the generation of customizable reports, and integrates with other databases.